AI-driven Platform for the generation of predictive tools from OMICs data
Empowering Biotech and Biomedical Research with a user-friendly, code-free AI solution to generate predictive tools.
About
The Digital Phenomics Platform is an online tool that enables users to build and use predictive tools for the classification of phenotypes using Machine Learning and Artificial intelligence on “omics” data. The tool enables users to upload their “omics” data, construct datasets, build predictive models, analyse models and generate predictions in a user-friendly and code-free manner. The Platform is organized into functional MODULES and its features, which are fully integrated and made available depending on their subscription plans.
The tool is currently in version beta 1.0, last updated on the 18th of August 2023. The Platform was developed and maintained by URA Informatics and Bioenhancer Systems. We invite all interested to test the main functionalities of the tool by subscribing to our demo version.
FEATURES
DATASET GENERATOR
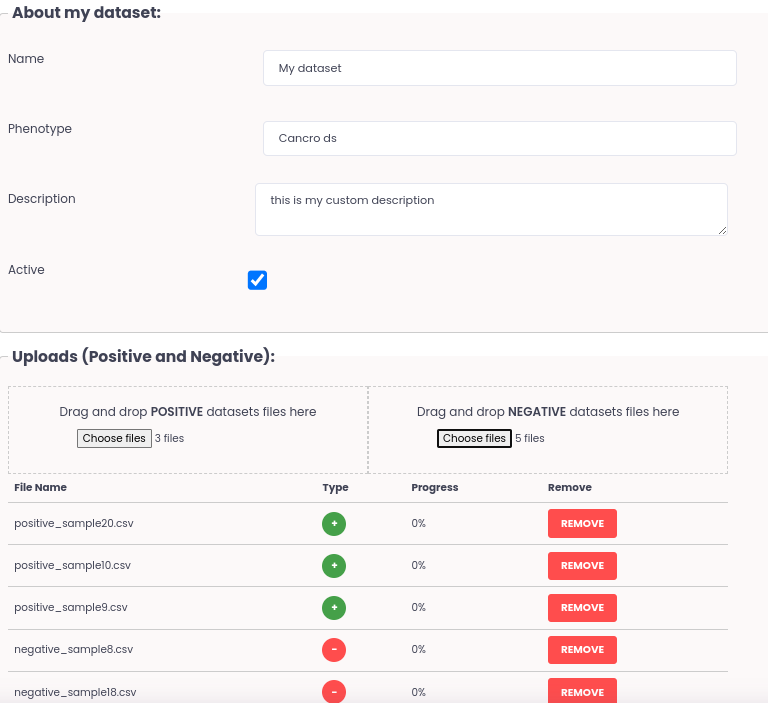
The Dataset generator is a platform feature included in the module GENERATOR. This feature enables the users to construct datasets with their omics data and associate them with a user-defined phenotype classification. Construction of datasets is simple, the users only need to drag and drop omics data files of individual samples to Positive or Negative to the phenotype and submit. Users can add and edit the information regarding the data/phenotype and save it to construct the dataset. The AI systems will then build and safely store an encrypted and binary file to be used downstream to other platform module features.
MODEL GENERATOR
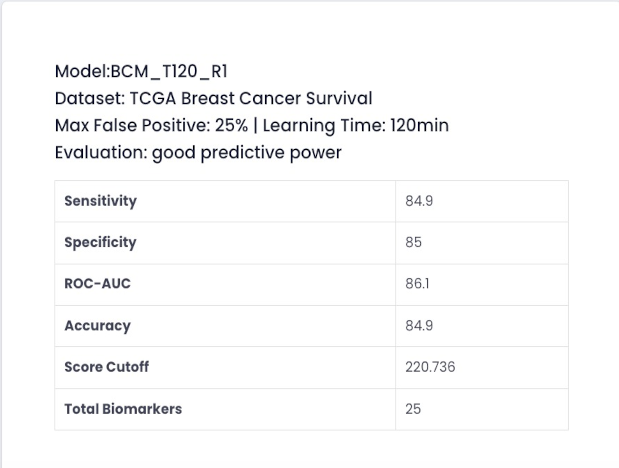
The Model generator is a platform feature included in the module GENERATOR. This feature enables users to build predictive models for the classification of phenotypes using previously constructed datasets. Model building is fully automated through our powerful AI engine (O2P-Mgen) upon saving your request. Users only need to choose the dataset and specify how much AI learning time is to be allocated and how much percentage of false positive rate is tolerable for your model. Once models are generated by AI, users can evaluate their models through the following visualization functionalities:
- Table with predictive biomarkers characteristics such as their median levels (phenotype positive), type of regulatory effect (e.g. up-regulation and down-regulation) and the associated p-values.
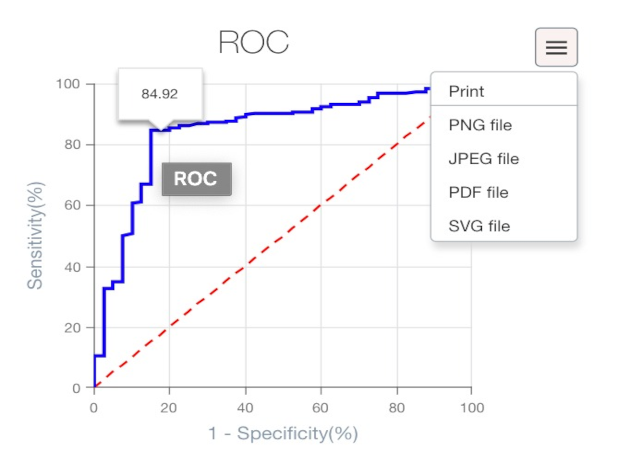
- Dynamic Receiver Operator Characteristic (ROC) curve, which can be downloaded as an Excel file or as a figure (multiple formats available);
- Table with multiple model performance metrics (sensitivity, specificity, accuracy, ROC-AUC, and predictive power auto evaluation);
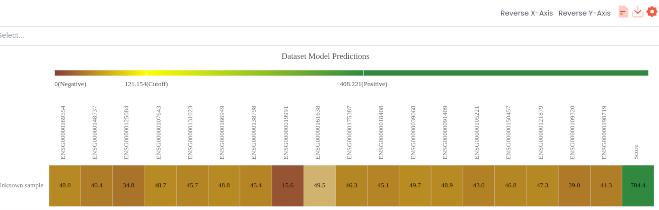
- Dynamic Heatmap with biomarkers partial scores and overall score on the dataset with auto-identification of false/true positive/negatives (can be downloaded and configurable);
Models are stored under encrypted and binary files which can be used downstream on other platform module features. Users can edit models and request new build attempts.
DATA PREDICTORS
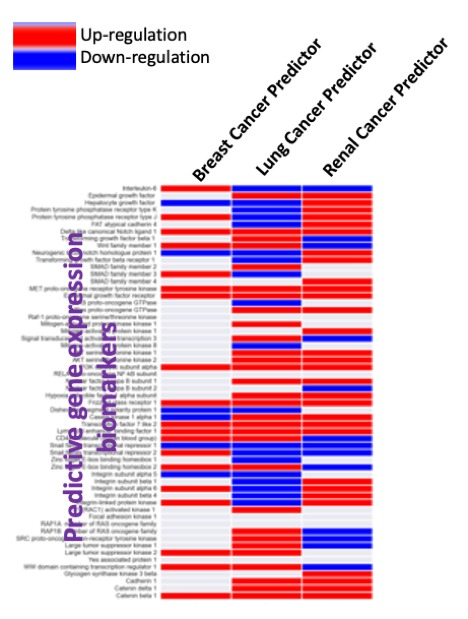
Digital Phenomics implements multiple mathematical models in our cutting-edge automated bioinformatic pipelines towards a phenomics analysis. We programme the pipelines for an automated and intelligent assessment of multiple phenotypes from large amounts of data coming from high-throughput technologies. This AI technology enables a rapid and effective checking of health status and screening of multiple diseases on data from the same biological sample. Human phenomics is an emerging field that focuses on the characterization of such traits on an individual (phenome). At DIGITAL Phenomics, we integrate high-throughput “omics” data from an individual towards characterizing the human disease phenome and providing a precision medicine insight. For this purpose, we often use scoring metrics for phenotype prediction and classification of diseases allowing us to build a phenome heatmap.
DEMO
ABOUT THE DEMO
Main platform features can be tested for free on our demo version. In this version, we included (built-in as default) tumour transcriptomic datasets for survival prognostic (the phenotype) of breast, lung and renal cancers. Users can build models (limited) using these datasets and test our powerful AI model generation tool. We also included initial models built to predict prognostics of breast, lung and renal cancers. Further, we made these models available to generate predictions on unknown tumour biopsies. To make predictions, users only need to add the values of the mRNA expression levels of the predictive biomarkers genes identified by the AI.
NOTE: Some functionalities may be limited or absent in this version

REGISTER FOR A DEMO ACCOUNT
To access our demo version, it is required to set up a demo account.
The demo account can be set by following the registration form by clicking the link below:
We only need from you the institution’s name, the user’s name and the institutional email.
Next, you will get a confirmation email to set your password securely;
Then, you can access the platform on its DEMO version through the LOGIN portal on the menu bar.
NOTE: Inactivity of accounts for over 6 months will be permanently deleted.
PLANS
FREE
£0
- Unlimited model generation runs
- Unlimited model predictions
- Built-in Datasets (only)
- AI-Learning time limited
- Limited models storage (up to 5)

Empowering Biotech and Biomedical Research with a user-friendly, code-free AI solution to generate predictive tools.
Contact us – [email protected]